What Conditions Would Need to Be Present in the Cell in Order to Reverse the Regulatory Conditions
Learning Objectives
- Compare inducible operons and repressible operons
- Draw why regulation of operons is of import
Each nucleated prison cell in a multicellular organism contains copies of the aforementioned DNA. Similarly, all cells in 2 pure bacterial cultures inoculated from the aforementioned starting colony contain the same Deoxyribonucleic acid, with the exception of changes that arise from spontaneous mutations. If each cell in a multicellular organism has the same DNA, then how is it that cells in different parts of the organism'southward body exhibit dissimilar characteristics? Similarly, how is it that the same bacterial cells within two pure cultures exposed to different environmental atmospheric condition can showroom different phenotypes? In both cases, each genetically identical cell does not turn on, or express, the same gear up of genes. Only a subset of proteins in a jail cell at a given fourth dimension is expressed.
Genomic Deoxyribonucleic acid contains both structural genes, which encode products that serve as cellular structures or enzymes, and regulatory genes, which encode products that regulate factor expression. The expression of a gene is a highly regulated process. Whereas regulating factor expression in multicellular organisms allows for cellular differentiation, in single-celled organisms similar prokaryotes, it primarily ensures that a cell'southward resources are not wasted making proteins that the cell does not need at that fourth dimension.
Elucidating the mechanisms controlling gene expression is important to the understanding of human health. Malfunctions in this process in humans lead to the evolution of cancer and other diseases. Understanding the interaction between the gene expression of a pathogen and that of its human host is important for the understanding of a particular infectious affliction. Gene regulation involves a complex web of interactions within a given cell among signals from the jail cell'south environment, signaling molecules within the cell, and the cell's Deoxyribonucleic acid. These interactions lead to the expression of some genes and the suppression of others, depending on circumstances.
Prokaryotes and eukaryotes share some similarities in their mechanisms to regulate gene expression; however, cistron expression in eukaryotes is more than complicated because of the temporal and spatial separation between the processes of transcription and translation. Thus, although most regulation of gene expression occurs through transcriptional control in prokaryotes, regulation of factor expression in eukaryotes occurs at the transcriptional level and postal service-transcriptionally (after the master transcript has been fabricated).
Prokaryotic Factor Regulation
In bacteria and archaea, structural proteins with related functions are usually encoded together within the genome in a block chosen an operon and are transcribed together under the control of a unmarried promoter, resulting in the formation of a polycistronic transcript (Figure ane). In this way, regulation of the transcription of all of the structural genes encoding the enzymes that catalyze the many steps in a single biochemical pathway tin be controlled simultaneously, considering they will either all be needed at the aforementioned time, or none will be needed. For case, in E. coli , all of the structural genes that encode enzymes needed to use lactose equally an energy source prevarication side by side to each other in the lactose (or lac) operon nether the control of a unmarried promoter, the lac promoter. French scientists François Jacob (1920–2013) and Jacques Monod at the Pasteur Institute were the start to show the organisation of bacterial genes into operons, through their studies on the lac operon of East. coli. For this work, they won the Nobel Prize in Physiology or Medicine in 1965. Although eukaryotic genes are not organized into operons, prokaryotic operons are excellent models for learning nigh factor regulation generally. There are some cistron clusters in eukaryotes that function like to operons. Many of the principles can exist applied to eukaryotic systems and contribute to our understanding of changes in gene expression in eukaryotes that can effect pathological changes such equally cancer.

Figure i. In prokaryotes, structural genes of related function are often organized together on the genome and transcribed together nether the control of a unmarried promoter. The operon's regulatory region includes both the promoter and the operator. If a repressor binds to the operator, then the structural genes will non be transcribed. Alternatively, activators may demark to the regulatory region, enhancing transcription.
Each operon includes DNA sequences that influence its own transcription; these are located in a region chosen the regulatory region. The regulatory region includes the promoter and the region surrounding the promoter, to which transcription factors, proteins encoded by regulatory genes, tin can demark. Transcription factors influence the binding of RNA polymerase to the promoter and allow its progression to transcribe structural genes. A repressor is a transcription factor that suppresses transcription of a gene in response to an external stimulus by binding to a Deoxyribonucleic acid sequence inside the regulatory region chosen the operator, which is located between the RNA polymerase binding site of the promoter and the transcriptional offset site of the first structural gene. Repressor binding physically blocks RNA polymerase from transcribing structural genes. Conversely, an activator is a transcription factor that increases the transcription of a factor in response to an external stimulus by facilitating RNA polymerase binding to the promoter. An inducer, a third type of regulatory molecule, is a small molecule that either activates or represses transcription by interacting with a repressor or an activator.
In prokaryotes, there are examples of operons whose gene products are required rather consistently and whose expression, therefore, is unregulated. Such operons are constitutively expressed, meaning they are transcribed and translated continuously to provide the cell with abiding intermediate levels of the protein products. Such genes encode enzymes involved in housekeeping functions required for cellular maintenance, including Dna replication, repair, and expression, as well as enzymes involved in core metabolism. In dissimilarity, there are other prokaryotic operons that are expressed only when needed and are regulated by repressors, activators, and inducers.
Recall almost It
- What are the parts in the DNA sequence of an operon?
- What types of regulatory molecules are at that place?
Regulation by Repression
Prokaryotic operons are normally controlled past the binding of repressors to operator regions, thereby preventing the transcription of the structural genes. Such operons are classified equally either repressible operons or inducible operons. Repressible operons, like the tryptophan (trp) operon, typically contain genes encoding enzymes required for a biosynthetic pathway. As long as the production of the pathway, like tryptophan, continues to be required past the jail cell, a repressible operon volition go along to exist expressed. However, when the product of the biosynthetic pathway begins to accumulate in the cell, removing the demand for the cell to proceed to brand more, the expression of the operon is repressed. Conversely, inducible operons, like the lac operon of E. coli , often incorporate genes encoding enzymes in a pathway involved in the metabolism of a specific substrate like lactose. These enzymes are only required when that substrate is available, thus expression of the operons is typically induced only in the presence of the substrate.
The trp Operon: A Repressible Operon
E. coli tin can synthesize tryptophan using enzymes that are encoded past 5 structural genes located next to each other in the trp operon (Effigy ii). When environmental tryptophan is low, the operon is turned on. This means that transcription is initiated, the genes are expressed, and tryptophan is synthesized. Nevertheless, if tryptophan is present in the surroundings, the trp operon is turned off. Transcription does not occur and tryptophan is not synthesized.
When tryptophan is not present in the cell, the repressor past itself does not bind to the operator; therefore, the operon is active and tryptophan is synthesized. Nonetheless, when tryptophan accumulates in the cell, 2 tryptophan molecules bind to the trp repressor molecule, which changes its shape, allowing it to bind to the trp operator. This bounden of the agile form of the trp repressor to the operator blocks RNA polymerase from transcribing the structural genes, stopping expression of the operon. Thus, the bodily production of the biosynthetic pathway controlled past the operon regulates the expression of the operon.

Effigy 2. The 5 structural genes needed to synthesize tryptophan in E. coli are located adjacent to each other in the trp operon. When tryptophan is absent, the repressor protein does non bind to the operator, and the genes are transcribed. When tryptophan is plentiful, tryptophan binds the repressor protein at the operator sequence. This physically blocks the RNA polymerase from transcribing the tryptophan biosynthesis genes.
Watch this video to learn more about the trp operon.
The lac Operon: An Inducible Operon
The lac operon is an example of an inducible operon that is also subject to activation in the absence of glucose (Figure 3). The lac operon encodes iii structural genes necessary to larn and process the disaccharide lactose from the environment, breaking it down into the elementary sugars glucose and galactose. For the lac operon to be expressed, lactose must be nowadays. This makes sense for the cell because it would exist energetically wasteful to create the enzymes to process lactose if lactose was not available.
In the absence of lactose, the lac repressor is leap to the operator region of the lac operon, physically preventing RNA polymerase from transcribing the structural genes. Even so, when lactose is present, the lactose inside the jail cell is converted to allolactose. Allolactose serves every bit an inducer molecule, binding to the repressor and changing its shape so that it is no longer able to bind to the operator Deoxyribonucleic acid. Removal of the repressor in the presence of lactose allows RNA polymerase to move through the operator region and begin transcription of the lac structural genes.
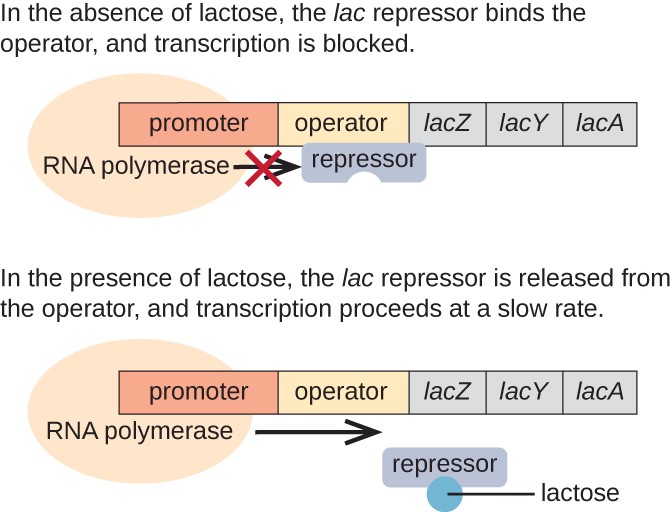
Effigy 3. The three structural genes that are needed to degrade lactose in E. coli are located next to each other in the lac operon. When lactose is absent, the repressor protein binds to the operator, physically blocking the RNA polymerase from transcribing the lac structural genes. When lactose is available, a lactose molecule binds the repressor poly peptide, preventing the repressor from binding to the operator sequence, and the genes are transcribed.
The lac Operon: Activation by Catabolite Activator Protein

Figure four. When grown in the presence of two substrates, E. coli uses the preferred substrate (in this case glucose) until it is depleted. Then, enzymes needed for the metabolism of the second substrate are expressed and growth resumes, although at a slower rate.
Bacteria typically have the ability to use a variety of substrates as carbon sources. Notwithstanding, because glucose is usually preferable to other substrates, bacteria take mechanisms to ensure that alternative substrates are just used when glucose has been depleted. Additionally, leaner have mechanisms to ensure that the genes encoding enzymes for using culling substrates are expressed only when the alternative substrate is available. In the 1940s, Jacques Monod was the first to demonstrate the preference for certain substrates over others through his studies of Eastward. coli'due south growth when cultured in the presence of 2 dissimilar substrates simultaneously. Such studies generated diauxic growth curves, like the ane shown in Figure 4. Although the preferred substrate glucose is used kickoff, Eastward. coli grows quickly and the enzymes for lactose metabolism are absent. However, once glucose levels are depleted, growth rates ho-hum, inducing the expression of the enzymes needed for the metabolism of the second substrate, lactose. Notice how the growth rate in lactose is slower, equally indicated by the lower steepness of the growth curve.
The power to switch from glucose apply to another substrate like lactose is a consequence of the activity of an enzyme called Enzyme IIA (EIIA). When glucose levels drop, cells produce less ATP from catabolism (see Catabolism of Carbohydrates), and EIIA becomes phosphorylated. Phosphorylated EIIA activates adenylyl cyclase, an enzyme that converts some of the remaining ATP to cyclic AMP (cAMP), a cyclic derivative of AMP and of import signaling molecule involved in glucose and energy metabolism in E. coli. Every bit a result, camp levels begin to rise in the cell (Figure five).

Effigy 5. When ATP levels decrease due to depletion of glucose, some remaining ATP is converted to camp by adenylyl cyclase. Thus, increased campsite levels signal glucose depletion.
The lac operon also plays a role in this switch from using glucose to using lactose. When glucose is scarce, the accumulating cAMP acquired by increased adenylyl cyclase activity binds to catabolite activator poly peptide (CAP), also known as army camp receptor poly peptide (CRP). The complex binds to the promoter region of the lac operon (Effigy 6). In the regulatory regions of these operons, a CAP bounden site is located upstream of the RNA polymerase binding site in the promoter. Binding of the CAP-camp complex to this site increases the binding ability of RNA polymerase to the promoter region to initiate the transcription of the structural genes. Thus, in the instance of the lac operon, for transcription to occur, lactose must be present (removing the lac repressor protein) and glucose levels must exist depleted (allowing binding of an activating protein). When glucose levels are high, in that location is catabolite repression of operons encoding enzymes for the metabolism of alternative substrates. Because of low cAMP levels under these weather condition, there is an insufficient amount of the CAP-cAMP complex to activate transcription of these operons. Run into Tabular array 1 for a summary of the regulation of the lac operon.

Effigy half dozen. (a) In the presence of army camp, CAP binds to the promoters of operons, like the lac operon, that encode genes for enzymes for the use of alternate substrates. (b) For the lac operon to exist expressed, in that location must be activation by cAMP-CAP equally well as removal of the lac repressor from the operator.
| Table 1. Conditions Affecting Transcription of the lac Operon | ||||
|---|---|---|---|---|
| Glucose | CAP binds | Lactose | Repressor binds | Transcription |
| + | – | – | + | No |
| + | – | + | – | Some |
| – | + | – | + | No |
| – | + | + | – | Yeah |
Picket an blithe tutorial about the workings of lac operon here.
Call back virtually It
- What affects the bounden of the trp operon repressor to the operator?
- How and when is the behavior of the lac repressor protein contradistinct?
- In addition to being repressible, how else is the lac operon regulated?
Global Responses of Prokaryotes
In prokaryotes, there are too several higher levels of gene regulation that have the ability to control the transcription of many related operons simultaneously in response to an environmental signal. A group of operons all controlled simultaneously is called a regulon.
Alarmones
When sensing impending stress, prokaryotes change the expression of a wide multifariousness of operons to answer in coordination. They do this through the production of alarmones, which are small intracellular nucleotide derivatives. Alarmones modify which genes are expressed and stimulate the expression of specific stress-response genes. The apply of alarmones to alter factor expression in response to stress appears to be of import in pathogenic bacteria. On encountering host defense mechanisms and other harsh conditions during infection, many operons encoding virulence genes are upregulated in response to alarmone signaling. Knowledge of these responses is primal to existence able to fully understand the infection procedure of many pathogens and to the development of therapies to counter this procedure.
Alternate σ Factors
Since the σ subunit of bacterial RNA polymerase confers specificity as to which promoters should be transcribed, altering the σ factor used is another way for leaner to quickly and globally change what regulons are transcribed at a given time. The σ factor recognizes sequences within a bacterial promoter, so different σ factors will each recognize slightly different promoter sequences. In this way, when the cell senses specific ecology conditions, it may respond by changing which σ factor information technology expresses, degrading the onetime 1 and producing a new i to transcribe the operons encoding genes whose products will be useful under the new environmental condition. For example, in sporulating bacteria of the genera Bacillus and Clostridium (which include many pathogens), a group of σ factors controls the expression of the many genes needed for sporulation in response to sporulation-stimulating signals.
Remember about Information technology
- What is the proper name given to a collection of operons that tin can be regulated as a group?
- What type of stimulus would trigger the transcription of a dissimilar σ cistron?
Additional Methods of Regulation in Bacteria: Attenuation and Riboswitches
Although most gene expression is regulated at the level of transcription initiation in prokaryotes, there are also mechanisms to control both the completion of transcription as well as translation concurrently. Since their discovery, these mechanisms accept been shown to control the completion of transcription and translation of many prokaryotic operons. Considering these mechanisms link the regulation of transcription and translation directly, they are specific to prokaryotes, because these processes are physically separated in eukaryotes.
One such regulatory organisation is attenuation, whereby secondary stem-loop structures formed within the 5' end of an mRNA being transcribed determine if transcription to consummate the synthesis of this mRNA volition occur and if this mRNA will exist used for translation. Across the transcriptional repression mechanism already discussed, attenuation also controls expression of the trp operon in E. coli (Figure 7). The trp operon regulatory region contains a leader sequence called trpL between the operator and the offset structural cistron, which has four stretches of RNA that can base pair with each other in different combinations. When a terminator stalk-loop forms, transcription terminates, releasing RNA polymerase from the mRNA. Nevertheless, when an antiterminator stem-loop forms, this prevents the formation of the terminator stalk-loop, so RNA polymerase tin transcribe the structural genes.
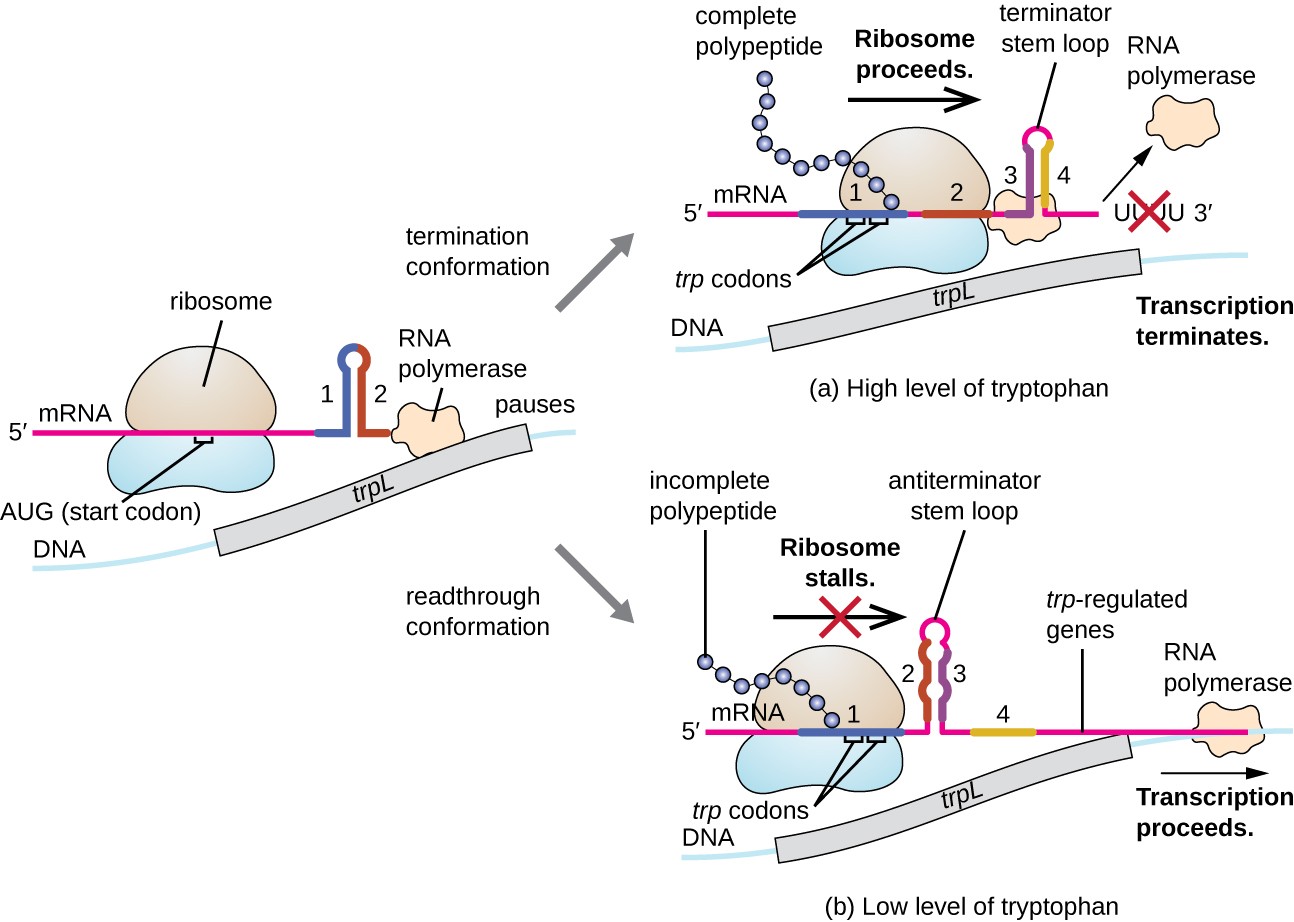
Figure seven. Click to view a larger image. When tryptophan is plentiful, translation of the short leader peptide encoded by trpL proceeds, the terminator loop between regions iii and 4 forms, and transcription terminates. When tryptophan levels are depleted, translation of the brusque leader peptide stalls at region 1, allowing regions 2 and 3 to form an antiterminator loop, and RNA polymerase tin transcribe the structural genes of the trp operon.
A related mechanism of concurrent regulation of transcription and translation in prokaryotes is the use of a riboswitch, a small region of noncoding RNA found within the 5' end of some prokaryotic mRNA molecules (Figure 8). A riboswitch may bind to a small intracellular molecule to stabilize certain secondary structures of the mRNA molecule. The binding of the small molecule determines which stem-loop structure forms, thus influencing the completion of mRNA synthesis and protein synthesis.

Figure 8. Click for a larger image. Riboswitches constitute inside prokaryotic mRNA molecules tin demark to small intracellular molecules, stabilizing sure RNA structures, influencing either the completion of the synthesis of the mRNA molecule itself (left) or the protein fabricated using that mRNA (right).
Other Factors Affecting Gene Expression in Prokaryotes and Eukaryotes
Although the focus on our discussion of transcriptional control used prokaryotic operons as examples, eukaryotic transcriptional control is similar in many ways. As in prokaryotes, eukaryotic transcription tin be controlled through the binding of transcription factors including repressors and activators. Interestingly, eukaryotic transcription can be influenced by the binding of proteins to regions of Dna, called enhancers, rather far away from the gene, through Dna looping facilitated between the enhancer and the promoter (Figure 9). Overall, regulating transcription is a highly constructive manner to command gene expression in both prokaryotes and eukaryotes. However, the control of cistron expression in eukaryotes in response to environmental and cellular stresses can exist achieved in additional ways without the bounden of transcription factors to regulatory regions.

Figure 9. In eukaryotes, an enhancer is a DNA sequence that promotes transcription. Each enhancer is made upwards of short Dna sequences chosen distal control elements. Activators bound to the distal control elements interact with mediator proteins and transcription factors. Two unlike genes may have the same promoter but different distal command elements, enabling differential gene expression.
Deoxyribonucleic acid-Level Control
In eukaryotes, the DNA molecules or associated histones can be chemically modified in such a way as to influence transcription; this is called epigenetic regulation. Methylation of certain cytosine nucleotides in Dna in response to environmental factors has been shown to influence apply of such Deoxyribonucleic acid for transcription, with DNA methylation commonly correlating to lowered levels of factor expression. Additionally, in response to environmental factors, histone proteins for packaging Deoxyribonucleic acid can besides be chemically modified in multiple ways, including acetylation and deacetylation, influencing the packaging state of Deoxyribonucleic acid and thus affecting the availability of loosely wound DNA for transcription. These chemic modifications can sometimes be maintained through multiple rounds of prison cell division, making at least some of these epigenetic changes heritable.
This video describes how epigenetic regulation controls gene expression.
Think about It
- What stops or allows transcription to keep when attenuation is operating?
- What determines the state of a riboswitch?
- Describe the function of an enhancer.
- Describe two mechanisms of epigenetic regulation in eukaryotes.
Clinical Focus: Travis, Resolution
This example concludes Travis's story that started in The Functions of Genetic Textile, RNA Transcription, and How Asexual Prokaryotes Achieve Genetic Diversity.
Although Travis survived his tour with necrotizing fasciitis, he would now have to undergo a skin-grafting surgery, followed past long-term concrete therapy. Based on the amount of muscle mass he lost, it is unlikely that his leg will return to full strength, but his physical therapist is optimistic that he will regain some use of his leg.
Laboratory testing revealed the causative agent of Travis's infection was a strain of group A streptococcus (Grouping A strep). As required by law, Travis's case was reported to the state health department and ultimately to the Centers for Disease Control and Prevention (CDC). At the CDC, the strain of grouping A strep isolated from Travis was analyzed more thoroughly for methicillin resistance.
Methicillin resistance is genetically encoded and is becoming more than common in group A strep through horizontal gene transfer. In necrotizing fasciitis, claret flow to the infected area is typically limited because of the action of various genetically encoded bacterial toxins. This is why at that place is typically little to no bleeding as a outcome of the incision test. Unfortunately, these bacterial toxins limit the effectiveness of intravenous antibiotics in clearing infection from the skin and underlying tissue, meaning that antibody resistance lone does not explain the ineffectiveness of Travis'southward handling. Nevertheless, intravenous antibiotic therapy was warranted to assistance minimize the possible outcome of sepsis, which is a common result of necrotizing fasciitis. Through genomic assay past the CDC of the strain isolated from Travis, several of the important virulence genes were shown to be encoded on prophages, indicating that transduction is important in the horizontal factor transfer of these genes from 1 bacterial cell to some other.
Key Concepts and Summary
- Factor expression is a tightly regulated process.
- Gene expression in prokaryotes is largely regulated at the point of transcription. Factor expression in eukaryotes is additionally regulated mail-transcriptionally.
- Prokaryotic structural genes of related function are frequently organized into operons, all controlled by transcription from a single promoter. The regulatory region of an operon includes the promoter itself and the region surrounding the promoter to which transcription factors can bind to influence transcription.
- Although some operons are constitutively expressed, most are subject to regulation through the use of transcription factors (repressors and activators). A repressor binds to an operator, a DNA sequence inside the regulatory region betwixt the RNA polymerase binding site in the promoter and first structural gene, thereby physically blocking transcription of these operons. An activator binds within the regulatory region of an operon, helping RNA polymerase bind to the promoter, thereby enhancing the transcription of this operon. An inducer influences transcription through interacting with a repressor or activator.
- The trp operon is a classic instance of a repressible operon. When tryptophan accumulates, tryptophan binds to a repressor, which then binds to the operator, preventing further transcription.
- The lac operon is a classic example an inducible operon. When lactose is present in the jail cell, it is converted to allolactose. Allolactose acts as an inducer, bounden to the repressor and preventing the repressor from binding to the operator. This allows transcription of the structural genes.
- The lac operon is also discipline to activation. When glucose levels are depleted, some cellular ATP is converted into camp, which binds to the catabolite activator protein (CAP). The camp-CAP complex activates transcription of the lac operon. When glucose levels are loftier, its presence prevents transcription of the lac operon and other operons past catabolite repression.
- Minor intracellular molecules chosen alarmones are made in response to various ecology stresses, allowing bacteria to control the transcription of a group of operons, called a regulon.
- Leaner take the ability to modify which σ factor of RNA polymerase they use in response to environmental weather condition to speedily and globally change which regulons are transcribed.
- Prokaryotes have regulatory mechanisms, including attenuation and the apply of riboswitches, to simultaneously command the completion of transcription and translation from that transcript. These mechanisms work through the formation of stem loops in the 5' finish of an mRNA molecule currently being synthesized.
- There are boosted points of regulation of cistron expression in prokaryotes and eukaryotes. In eukaryotes, epigenetic regulation by chemical modification of DNA or histones, and regulation of RNA processing are two methods.
Multiple Pick
An operon of genes encoding enzymes in a biosynthetic pathway is likely to be which of the post-obit?
- inducible
- repressible
- constitutive
- monocistronic
Evidence Reply
Respond b. An operon of genes encoding enzymes in a biosynthetic pathway is probable to be repressible.
An operon encoding genes that are transcribed and translated continuously to provide the cell with constant intermediate levels of the protein products is said to be which of the following?
- repressible
- inducible
- constitutive
- activated
Show Answer
Answer c. This type of operon is said to be constitutive.
Which of the following conditions leads to maximal expression of the lac operon?
- lactose present, glucose absent
- lactose present, glucose present
- lactose absent, glucose absent
- lactose absent, glucose present
Show Answer
Answer a. Lactose present and glucose absent leads to maximal expression of the lac operon.
Which of the following is a blazon of regulation of gene expression unique to eukaryotes?
- attenuation
- utilise of alternate σ factor
- chemical modification of histones
- alarmones
Show Reply
Answer c. Chemical modification of histones is a type of regulation of gene expression unique to eukaryotes.
Fill up in the Blank
The Dna sequence, to which repressors may bind, that lies between the promoter and the first structural gene is called the ________.
Show Answer
The Dna sequence, to which repressors may bind, that lies between the promoter and the commencement structural gene is chosen the operator.
The prevention of expression of operons encoding substrate use pathways for substrates other than glucose when glucose is present is called _______.
Prove Answer
The prevention of expression of operons encoding substrate use pathways for substrates other than glucose when glucose is present is called catabolite repression.
Think about Information technology
- What are two ways that leaner can influence the transcription of multiple unlike operons simultaneously in response to a particular environmental condition?
- The post-obit figure is from Monod'due south original work on diauxic growth showing the growth of E. coli in the simultaneous presence of xylose and glucose every bit the just carbon sources. Explain what is happening at points A–D with respect to the carbon source existence used for growth, and explicate whether the xylose-apply operon is being expressed (and why). Annotation that expression of the enzymes required for xylose use is regulated in a manner similar to the expression of the enzymes required for lactose utilise.

Source: https://courses.lumenlearning.com/microbiology/chapter/gene-regulation-operon-theory/
0 Response to "What Conditions Would Need to Be Present in the Cell in Order to Reverse the Regulatory Conditions"
Post a Comment